

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
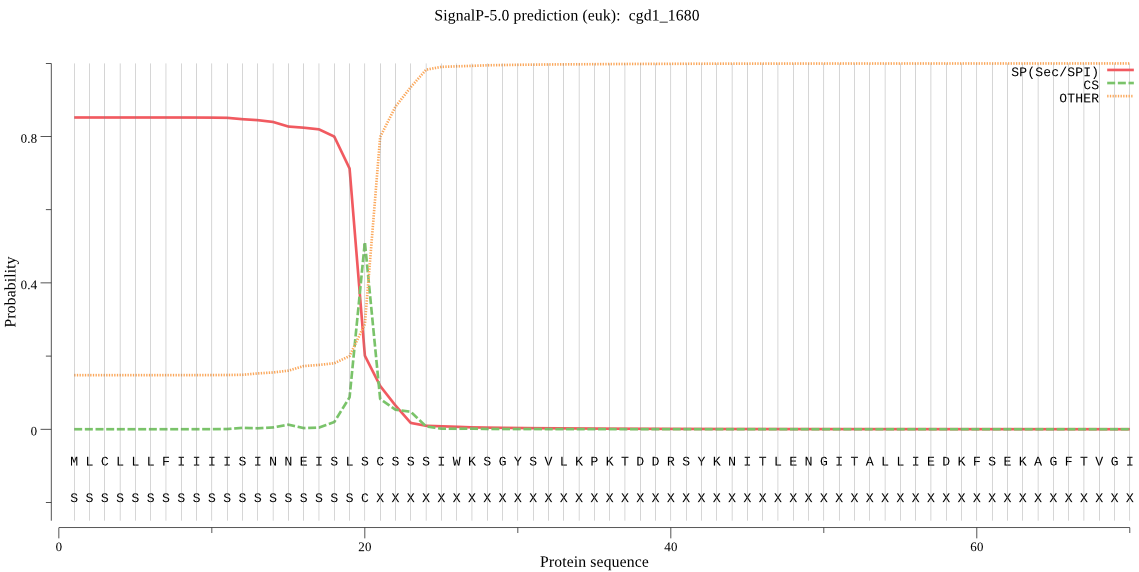
| cgd1_1680 | SP | 0.046456 | 0.953099 | 0.000445 | CS pos: 20-21. SLS-CS. Pr: 0.5020 |
MLCLLLFIIIISINNEISLSCSSSIWKSGYSVLKPKTDDRSYKNITLENGITALLIEDKF SEKAGFTVGIKVGSFNNPVYALGLFHLIEHVLFLGTKKYPAPESYDEFMAQHGGKNNAYT SEERTIYFNEIGEEYLEEGLDRFSHFFIDPLFYENVIEKEIHIINSEHLKNIPNEFDRLF HMLKTHTNKPMSQFTTGNIETLVDIPNKLGISIPRLLKKMYKKYYCGINMFIVLSSKRSL TDQEKLLQKYFSGVLIDNDGQCEFSSLKKEHGILNKSIIDQKYLSKKIHVKSLGGRDLLW LIWSFPARLISPVKQPLIYLSYILNSKQKNSLFWFLQKNNYITNSNSVYENYTFGSIFIY QLELTSEGLKNQFEIIGLIYKYINKLKESKELLKVYQGIRSLTEREFITNTEMLESSPMH STSEICSKMIQYGVHAALSGDILIEDVDENLIYEILNAISPFDTLFLVSDEQEFSGTYEK FFHVKHAIEDIPIKTLNAWKKTKFNEREENEIKLPTPEKCSPINLRIIQEVEDLSTPQRL DSMLANIWWNGPVKKSHKIGIKILLKFPRRYNKGIETQVWGEIITYILDTLIKEKIERYS ECGMSFYIEWDVEGILISIDTFGYSDDIDILLNEIVASEIANISNFDCSILNEIIAELNN SKYAFNSEDTTYSKIMLIIKSLQTNGEYTEWEYRNFINRMFVEIENQNPSKNRHKFISFI LEYTQTILGSSSNKGYNKCELFNSWAYKLLHRQSIIAYLQGNISKNKSLYLIEKFVLNSK ILSLNDKYSMKKKIHKLTRPIDIALINPVFEDINNTVLAFYQFGVPSFEEKLHLMALQPI IQGYIYDNLRTNKQLGYIIFANIVPISSTRLLVVGVEGDNNNSVEKIESIIRNTLYEFST RKLGNMESHMFEDIKSALIQEAKSIGNSFNQKLNHYWDEIRYVGDLSESFNLQRAIDYIN NKMTIEHLYNTFSKLINSKERTRSHSTIKAIYYSKTSPEFKKQEFIDKYMNSALSIARMS LKEDDFY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd1_1680 | 121 S | NAYTSEERT | 0.993 | unsp | cgd1_1680 | 121 S | NAYTSEERT | 0.993 | unsp | cgd1_1680 | 121 S | NAYTSEERT | 0.993 | unsp | cgd1_1680 | 239 S | SSKRSLTDQ | 0.996 | unsp | cgd1_1680 | 277 S | ILNKSIIDQ | 0.994 | unsp | cgd1_1680 | 285 S | QKYLSKKIH | 0.992 | unsp | cgd1_1680 | 403 T | IRSLTEREF | 0.993 | unsp | cgd1_1680 | 421 S | SPMHSTSEI | 0.994 | unsp | cgd1_1680 | 460 S | LNAISPFDT | 0.994 | unsp | cgd1_1680 | 924 S | QEAKSIGNS | 0.994 | unsp | cgd1_1680 | 978 S | KLINSKERT | 0.993 | unsp | cgd1_1680 | 986 S | TRSHSTIKA | 0.997 | unsp | cgd1_1680 | 997 S | YSKTSPEFK | 0.996 | unsp | cgd1_1680 | 1020 S | IARMSLKED | 0.998 | unsp | cgd1_1680 | 41 S | TDDRSYKNI | 0.996 | unsp | cgd1_1680 | 61 S | EDKFSEKAG | 0.996 | unsp |