

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
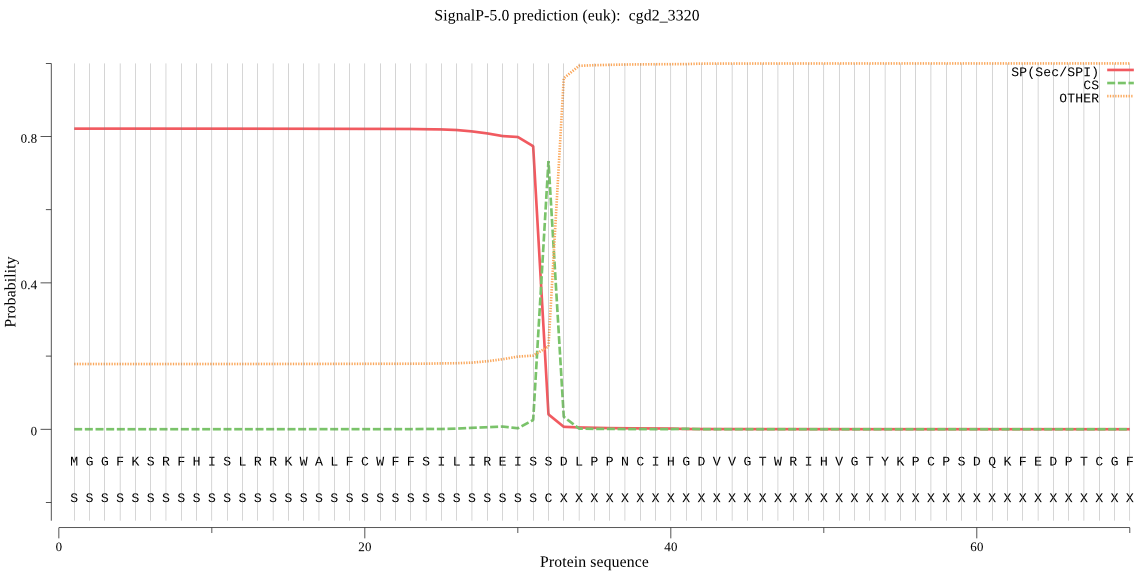
| cgd2_3320 | SP | 0.022690 | 0.976112 | 0.001198 | CS pos: 32-33. ISS-DL. Pr: 0.9618 |
MGGFKSRFHISLRRKWALFCWFFSILIREISSDLPPNCIHGDVVGTWRIHVGTYKPCPSD QKFEDPTCGFPSPDRDFAHSMLTPTERGLMNNNFKLSFTFDVKFEGSELKVISTQNLDGV DTGKFNDFEKVGTTGNWTVILDQAFTFWTKSYRYTAFFKYINEHEDINASYCYCHTTLLG WWDSYPESVISNQDNKEENKILSLEERSKYNNYPWLVMSEDLKLRRGCWYGMRIFDGDGK MTDRTEWTLKLPRWRSSPLGLPPDHPVNAQNPTIAEYLNEVSAKYSEFESKNKLWSRSNK FEIGDIQNLDTLPKVRKALKISPHFVQRKKSNDEYITEISSVREEEQISGSKNNTDEPIW MRIRSFDWANPQHVYGRLGKKVRLIPEVFNQGDCGDCFAVTAATIITSRLWIKYSNEPDI LRKIYASSIQMSECNVYNQGCGGGLITLAFKFAQEVGVRTQDCVDDYAKNLGLKEIYPTP VYTPDEVGIADDGHSFLQTKKGKKFESKKSKIKEIKQQEGDNQEYDQEHDQEHDQEHEQE HEQEHEQEHEQEHEREHDQEQGQKHDQKYQDDYKDMEEHLEGSDSAEHISNVHDYYGDSD DENSSDNEELGHLSGAKYNIPKKFNVIQQLCWDLGGHMGAANTQCRKDIPITKNIPKSCS KIIKVKEYSYVNNVYGKTTARDIMESLWNEGPVAVSLEPTLEFSLYNSGVFKGFYDPITR QYPWSSIPWHKVDHAMVITGWGWETYGNERIPYWIVQNSWGKRWGEKGFCRIIRGVNELS IEHAAVRASVKIYDSDRKYKAADLQKVHDESVFQYL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd2_3320 | 789 S | AVRASVKIY | 0.993 | unsp | cgd2_3320 | 789 S | AVRASVKIY | 0.993 | unsp | cgd2_3320 | 789 S | AVRASVKIY | 0.993 | unsp | cgd2_3320 | 331 S | QRKKSNDEY | 0.993 | unsp | cgd2_3320 | 341 S | TEISSVREE | 0.997 | unsp |