

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| cgd8_600 | SP | 0.027435 | 0.972329 | 0.000236 | CS pos: 23-24. VYG-EE. Pr: 0.9066 |
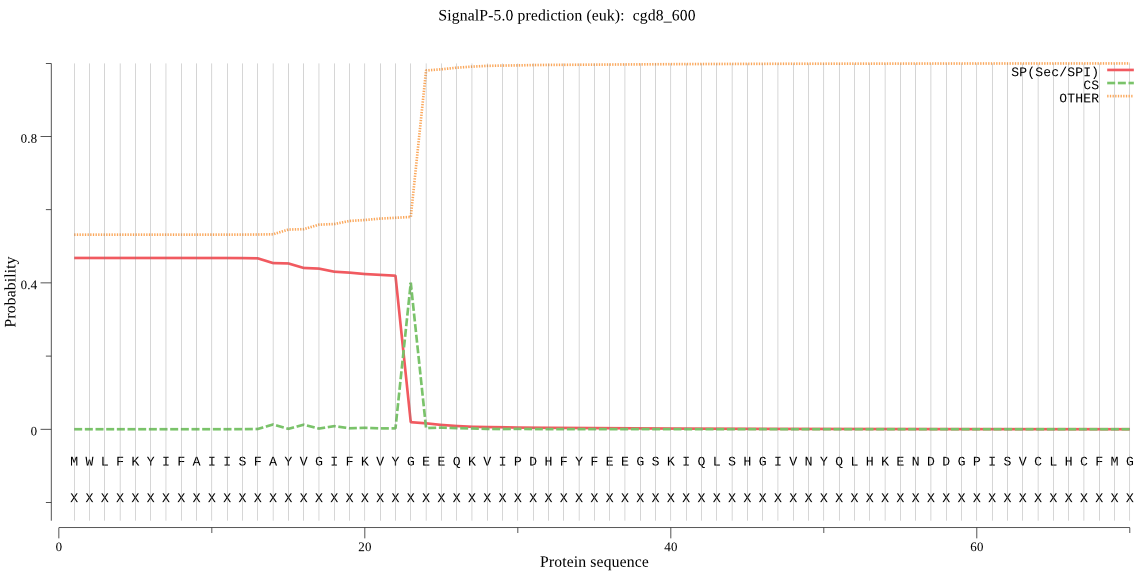
MWLFKYIFAIISFAYVGIFKVYGEEQKVIPDHFYFEEGSKIQLSHGIVNYQLHKENDDGP ISVCLHCFMGTISDCSSISKNLAKNGYRALRFDFYGHGLSQYKNFGQYSVDDYVDQTMEL LEKLGLYNVTAISEEELHSSSFIPKLHVIGTSLGGFVAMRIAQRFPKHISKLVLDAPPGL LKKKVAPYLQYSIINYPLQLFANIYSPVWGCYQFMDPPSENLSSPKLDFRAKHYCKQILL TSLQLFLGINLWNNQQIYQEFSKVDVPTLFFWGAEDRLCPLSSAITILNEYVPNTKIIVY ENCKHRCSKYCKEQFVEDVIKYFNNELDQELVSMSQYYNSVHDIVHSSEKRSLPSIRGTK TDTLLEPSSIPEDTCSTSTSGSQEIAVQLASSTEENTETGSSMSSSENFDDFCKTKN
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd8_600 | T | 379 | 0.563 | 0.077 | cgd8_600 | T | 393 | 0.553 | 0.054 | cgd8_600 | T | 377 | 0.552 | 0.218 | cgd8_600 | T | 363 | 0.537 | 0.018 | cgd8_600 | T | 374 | 0.537 | 0.032 | cgd8_600 | T | 359 | 0.530 | 0.035 | cgd8_600 | T | 361 | 0.530 | 0.056 | cgd8_600 | S | 391 | 0.519 | 0.029 | cgd8_600 | S | 369 | 0.504 | 0.067 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd8_600 | T | 379 | 0.563 | 0.077 | cgd8_600 | T | 393 | 0.553 | 0.054 | cgd8_600 | T | 377 | 0.552 | 0.218 | cgd8_600 | T | 363 | 0.537 | 0.018 | cgd8_600 | T | 374 | 0.537 | 0.032 | cgd8_600 | T | 359 | 0.530 | 0.035 | cgd8_600 | T | 361 | 0.530 | 0.056 | cgd8_600 | S | 391 | 0.519 | 0.029 | cgd8_600 | S | 369 | 0.504 | 0.067 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cgd8_600 | 392 S | QLASSTEEN | 0.996 | unsp | cgd8_600 | 392 S | QLASSTEEN | 0.996 | unsp | cgd8_600 | 392 S | QLASSTEEN | 0.996 | unsp | cgd8_600 | 402 S | ETGSSMSSS | 0.992 | unsp | cgd8_600 | 404 S | GSSMSSSEN | 0.996 | unsp | cgd8_600 | 405 S | SSMSSSENF | 0.993 | unsp | cgd8_600 | 133 S | VTAISEEEL | 0.99 | unsp | cgd8_600 | 355 S | RSLPSIRGT | 0.993 | unsp |