◉ The Genomic SSRs Millets DataBase (GSMDB) interface is organized into seven distinct tabs: Home, SSRs, BLAST, JBrowse, Statistics, Tutorial, and Contact. Each tab serves a specific function to facilitate efficient navigation and utilization of the database. The 'Home' section serves as an introduction to the database, providing comprehensive details about simple sequence repeats (SSRs) and millet
◉ In the 'SSRs' tab, users can delve into SSR data by choosing their preferred millet species. This section acts as the central hub for accessing and refining queries, allowing users to specify various parameters, such as chromosome/scaffold/contig, repeat type, and genomic region, once a specific species is selected (Figure 1)

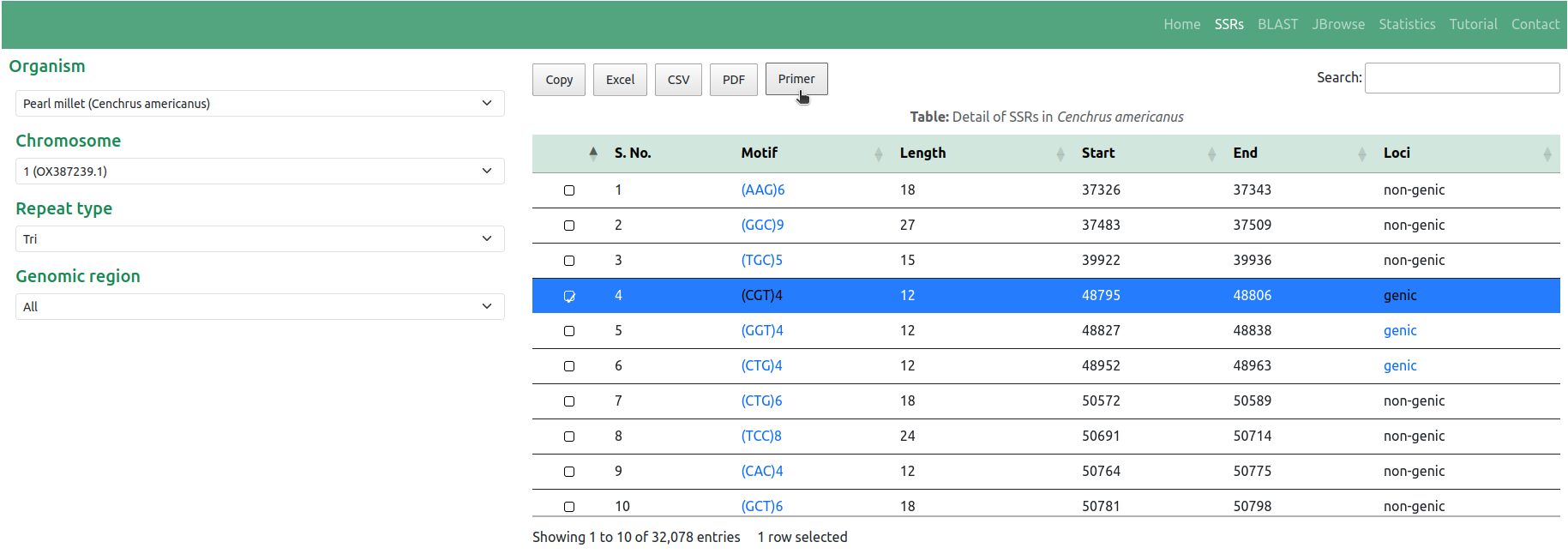
◉ This feature enables users to retrieve SSR details presented in a versatile tabular format. The tabular representation includes essential information such as Motif, Length, Start, End, and Loci details, all aligned with the specific selection criteria chosen by the user. Additionally, users have the option to search for specific motifs within the table using the integrated search bar. Furthermore, the interface offers options to export the table content in various formats through respective buttons, thereby enhancing data usability (Figure 2)
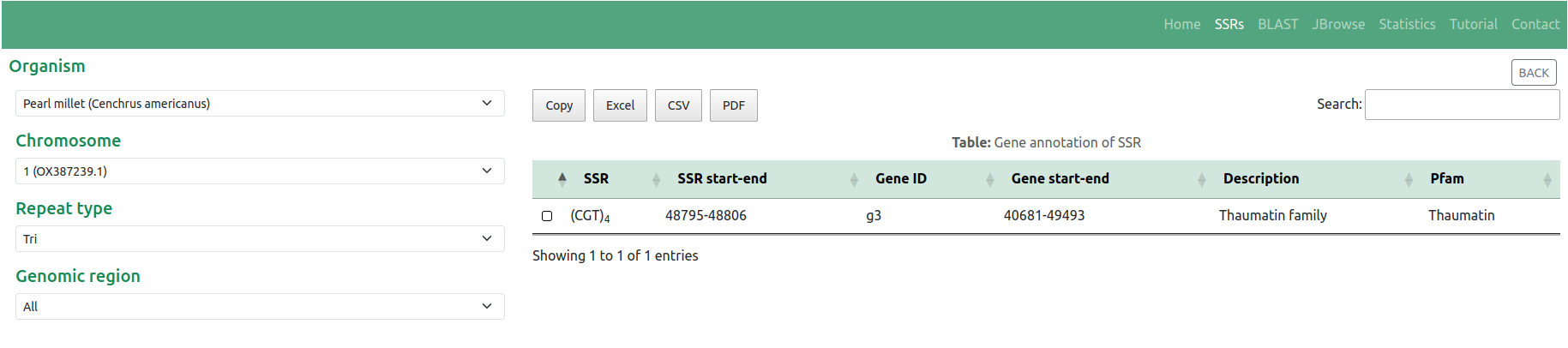
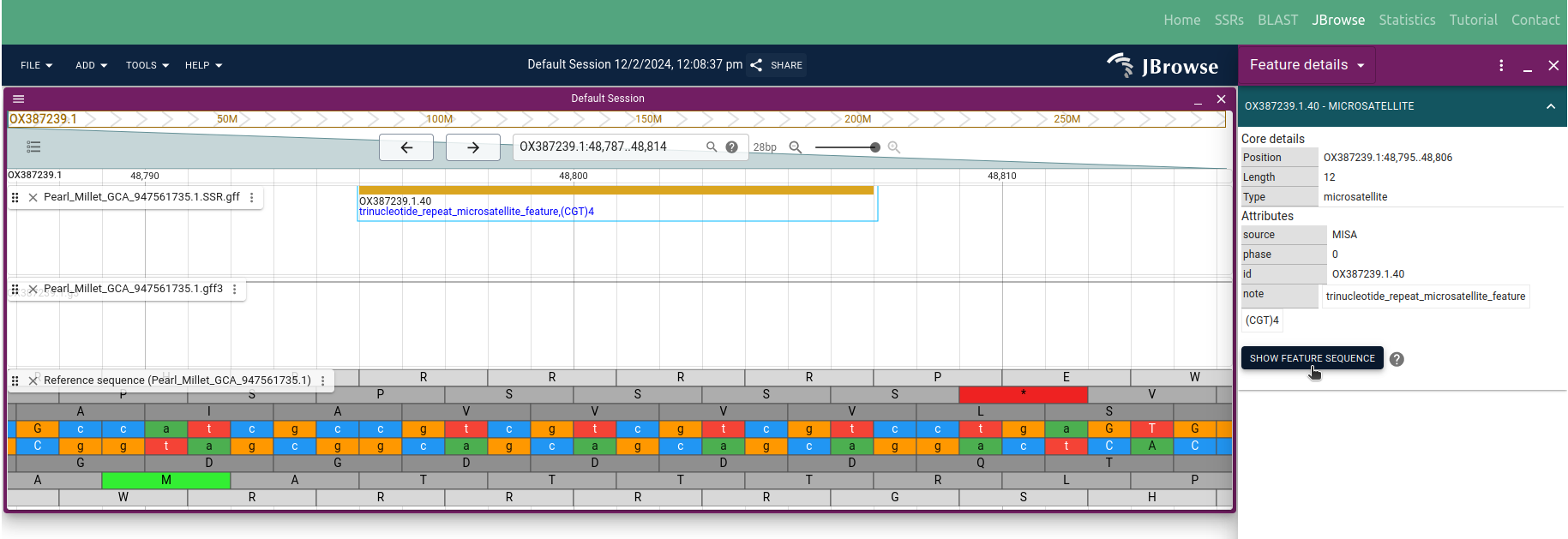
Users can access gene annotations by clicking on the respective 'Loci' in the table (Figure 3). Similarly, users can seamlessly navigate to the integrated JBrowse to visualize SSRs on the respective genome sequence by clicking on the desired 'Motif' in the table. For example, users can examine the SSR '(CGT)4' with Start: 48795 & End: 48806 position as depicted in Figure 11
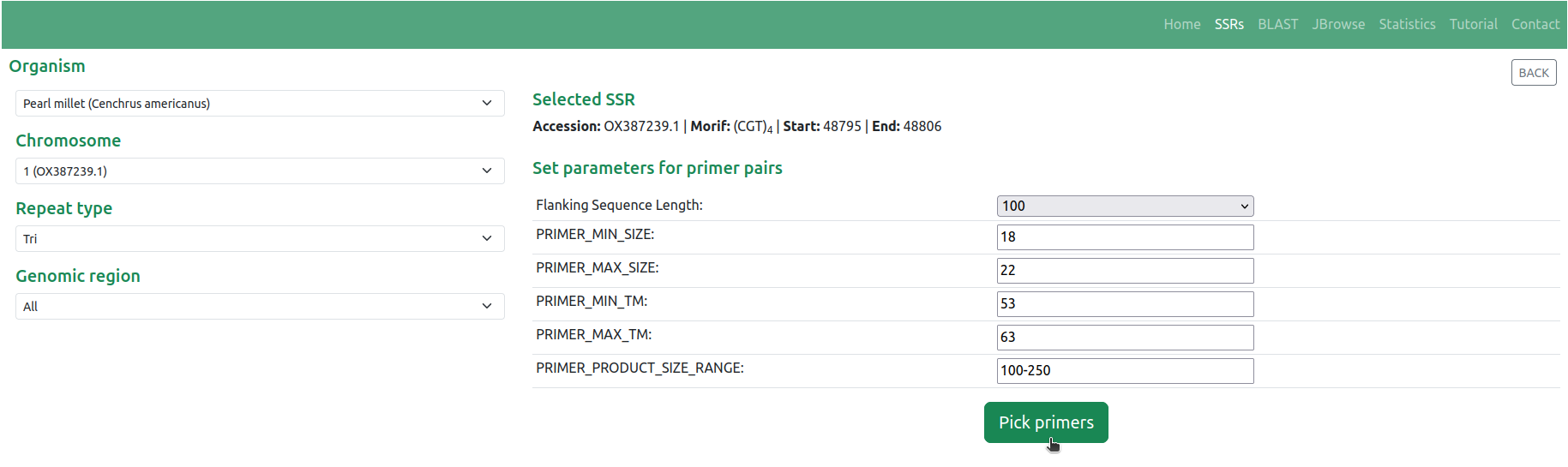
◉ Furthermore, the database offers the functionality of designing primer pairs targeting the SSRs using the Primer3 tool. Notably, the table includes a 'Primer' button (Figure 2), enabling users to design primer pairs by selecting the SSR of interest. This 'Primer' button serves as a pivotal tool, allowing users to customize the flanking sequence length and other fundamental parameters necessary for primer design for the selected SSR. Once these parameters are specified, users can obtain the results of the designed primer pairs by activating the 'Pick primers' button (Figure 4)
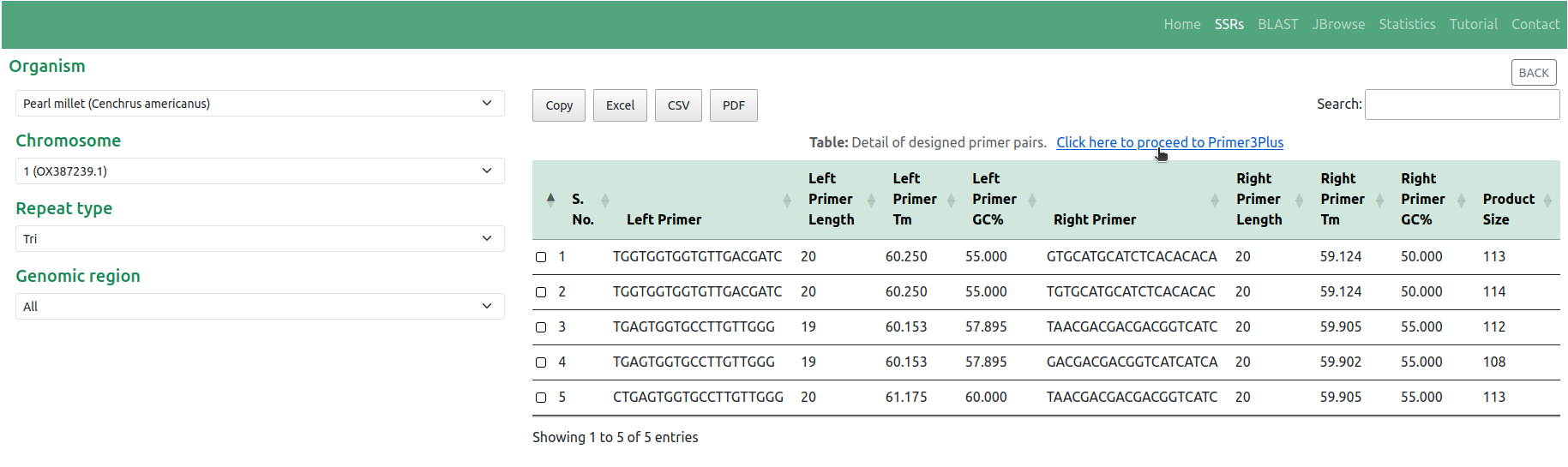
◉ This action prompts the display of comprehensive details regarding the designed primer pairs. For users interested in utilizing advanced parameters in their primer design, they have the option to proceed to the Primer3Plus web page with their selected SSR and flanking sequence by clicking on the link 'Click here to proceed to Primer3Plus' (Figure 5)
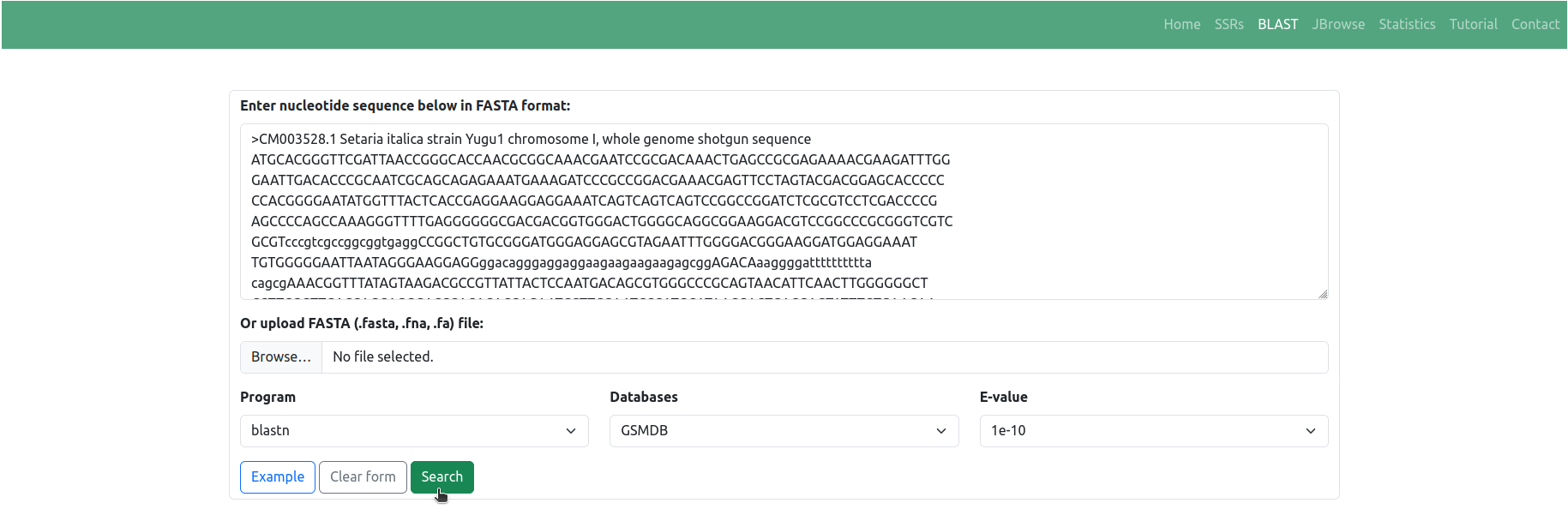
◉ The 'BLAST' tab facilitates a sequence-based search of SSRs within the GSMDB. Users can input nucleotide sequences in FASTA format directly into the provided textarea, or alternatively, they have the option to upload a file (.fasta, .fna, .fa) containing nucleotide sequences for analysis. Upon clicking the 'Search' button, the BLAST (blastn) algorithm executes a sequence similarity search against the comprehensive collection of millet sequences cataloged within the GSMDB (Figure 6)
◉ The search results are displayed in a user-friendly tabular format, outlining matches between the query and genome sequences of millets available in the GSMDB. These matches are based on alignment scores and sequence identities. Users can conveniently review the identified SSRs along with pertinent information such as SSR motif, genomic location, and associated annotations by clicking on the respective 'Subject ID' in the table (Figure 7)
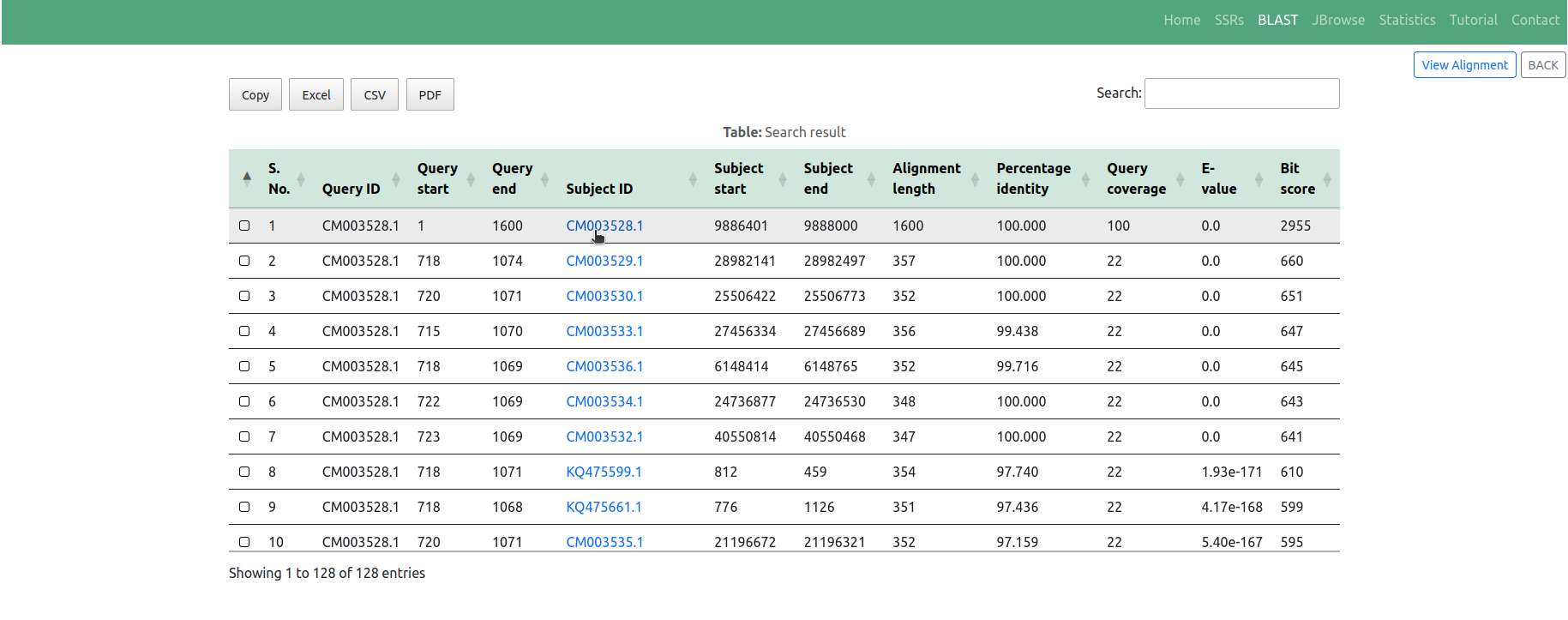
◉ Additionally, users have the option to visualize sequence similarity results by clicking on the 'View Alignment' button (Figure 7). This feature offers a visually appealing, customizable, and highly interactive presentation of aligned sequences along with their corresponding scores, facilitating interactive and intuitive navigation of the alignment results (Figure 8)

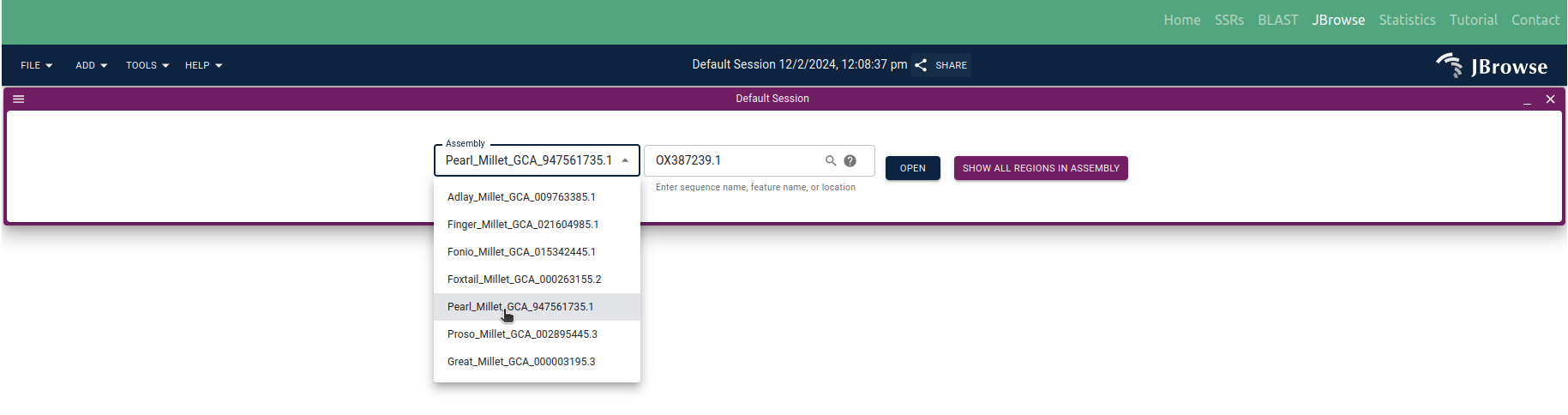
◉ The 'JBrowse' tab facilitates the visualization of identified SSRs locations on their respective genome sequences. Here, users can 'OPEN' the organism of interest and the respective chromosome/scaffold/contig using the dropdown menu (Figure 9)
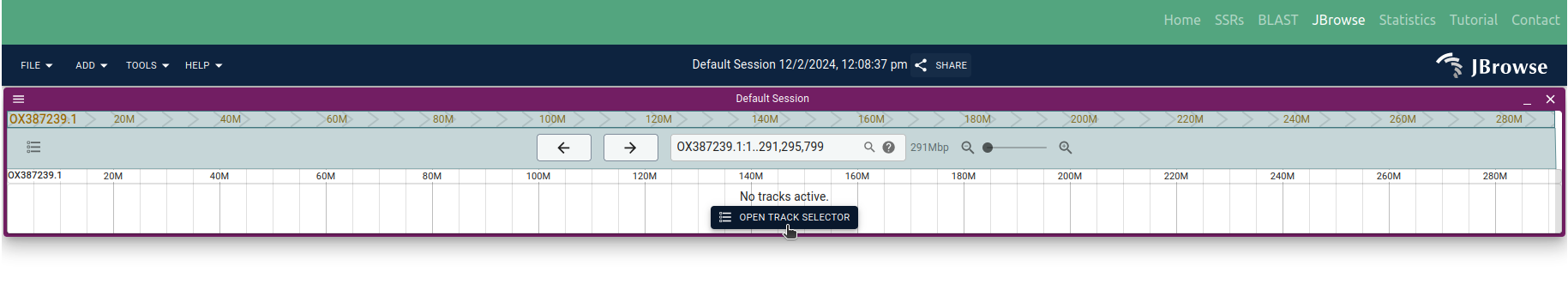
◉ Once the organism is selected, users can further select from available tracks, including the Reference genome, SSRs, and Feature track using 'OPEN TRACK SELECTOR'(Figure 10)
◉ Following the track selection, users have the ability to define the range of the genome sequence they wish to visualize, allowing them to focus on specific regions of interest. This functionality enables users to precisely examine SSRs within the selected area, enhancing the precision and efficiency of the visualization process. See example SSR '(CGT)4' with Start: 48795 & End: 48806 in JBrowse as shown in the Figure 11; and for additional functionalities, please refer to the JBrowse user guide
◉ The 'Statistic' tab offers a graphical representation of the total SSRs accessible within the GSMDB. In contrast, the 'Tutorial' page within the database explicates the functionality and interpretation of the available data. Additionally, the database includes a 'Contact' tab that offers information about GSMDB developers, and users have the ability to convey their inquiries or suggestions to the developers via the this tab.
***